 training data
training data 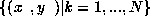 ,
we can approximate
,
we can approximate
Parzen windows are commonly used to approximate densities. Given
 training data
training data 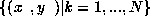 ,
we can approximate
,
we can approximate
where
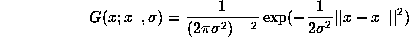
is a multidimensional properly normalized
Gaussian centered at data  with
variance
with
variance 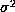 . It has been shown
(Duda and Hart (1973)) that Parzen windows approximate densities
for
. It has been shown
(Duda and Hart (1973)) that Parzen windows approximate densities
for  arbitrarily well, if
arbitrarily well, if  is appropriately
scaled.
is appropriately
scaled.
Using Parzen windows we may write
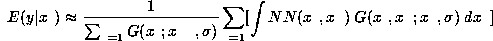
where we have used the fact that

and where  is a Gaussian projected onto
the known input dimensions (by simply leaving out the unknown dimensions
in the exponent and in the normalization, see Ahmad and Tresp, 1993).
is a Gaussian projected onto
the known input dimensions (by simply leaving out the unknown dimensions
in the exponent and in the normalization, see Ahmad and Tresp, 1993).
 are the components of the training data corresponding
to the known input (compare Figure 1).
are the components of the training data corresponding
to the known input (compare Figure 1).
Now, if we assume that the network prediction is approximately
constant over the ``width'' of the Gaussians,  ,
we can approximate
,
we can approximate

where  is the network prediction
which we obtain if we substituted the corresponding components of the
training data for the unknown inputs.
is the network prediction
which we obtain if we substituted the corresponding components of the
training data for the unknown inputs.
With this approximation,
Interestingly, we have obtained a network of normalized Gaussians which
are centered at the known components of the
data points. The ''output weights''  consist of the neural network predictions where for the unknown input
the corresponding components of the training data points have been
substituted.
Note, that we have obtained an approximation which has the same structure
as the solution for normalized Gaussian basis functions
(Ahmad and Tresp, 1994).
consist of the neural network predictions where for the unknown input
the corresponding components of the training data points have been
substituted.
Note, that we have obtained an approximation which has the same structure
as the solution for normalized Gaussian basis functions
(Ahmad and Tresp, 1994).
In many applications it might be easy to select a reasonable value for
 using prior knowledge but there are also two simple ways
to obtain a good estimate for
using prior knowledge but there are also two simple ways
to obtain a good estimate for  using leave-one-out methods.
The first method consists of removing
the
using leave-one-out methods.
The first method consists of removing
the  pattern from the training data and calculating
pattern from the training data and calculating
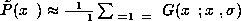 .
Then select the
.
Then select the  for which the log likelihood
for which the log likelihood
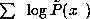 is maximum. The second method consists
of treating an input of the
is maximum. The second method consists
of treating an input of the  training pattern as missing
and then testing how well our algorithm (Equation 2) can predict the target.
Select the
training pattern as missing
and then testing how well our algorithm (Equation 2) can predict the target.
Select the  which gives the best performance.
In this way it would even be possible to select
input-dimension-specific widths
which gives the best performance.
In this way it would even be possible to select
input-dimension-specific widths  leading to ``elliptical'', axis-parallel Gaussians (Ahmad and Tresp, 1993).
leading to ``elliptical'', axis-parallel Gaussians (Ahmad and Tresp, 1993).
Note that the complexity of the solution is independent of the number of missing inputs! In contrast, the complexity of the solution for feedforward networks suggested in Tresp, Ahmad and Neuneier (1994) grows exponentially with the number of missing inputs. Although similar in character to the solution for normalized RBFs, here we have no restrictions on the network architecture which allows us to choose the network most appropriate for the application.
If the amount of training data is large, one can use the following approximations:
 nearest data points. The distance
is determined based on the known inputs.
nearest data points. The distance
is determined based on the known inputs. 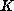 can
probably be reasonably small
can
probably be reasonably small  . In the extreme case,
. In the extreme case,
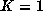 and we obtain a nearest-neighbor solution.
Efficient tree-based algorithms exist for computing the
and we obtain a nearest-neighbor solution.
Efficient tree-based algorithms exist for computing the
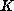 -nearest neighbors.
-nearest neighbors.
Note that the solution which substitutes the components of the training data closest to the input seems biologically plausible.